Roi select: Difference between revisions
No edit summary |
mNo edit summary |
||
| Line 1: | Line 1: | ||
{{DISPLAYTITLE:roi_select}} | |||
<span style="color:red">{{infobox|This page is incomplete and currently under development.}}</span> | <span style="color:red">{{infobox|This page is incomplete and currently under development.}}</span> | ||
| Line 5: | Line 6: | ||
=Introduction= | =Introduction= | ||
After we run DTI data through a pipeline (whether [[Diffusion Processing Pipeline|our in-house one]] or a third-party one, such as [http://www.trackvis.org/dtk/ DTK]), we are left with data for hundreds of thousands of streamlines. Often times, however, we are interested in only a small subset of that data. We may be working with a specific structure in the brain and would like to see a visualization of only that structure. Or we may be interested in a particular region and would like to compute statistics about the streamlines that pass through that region. Perhaps we want to see how many streamlines connect two distinct regions and what paths they take. The <tt>roi_select</tt> tool can help achieve these goals. | After we run DTI data through a pipeline (whether [[Diffusion Processing Pipeline|our in-house one]] or a third-party one, such as [http://www.trackvis.org/dtk/ DTK]), we are left with data for hundreds of thousands of streamlines. Often times, however, we are interested in only a small subset of that data. We may be working with a specific structure in the brain and would like to see a visualization of only that structure. Or we may be interested in a particular region and would like to compute statistics about the streamlines that pass through that region. Perhaps we want to see how many streamlines connect two distinct regions and what paths they take. The <tt>roi_select</tt> tool can help achieve these goals. | ||
=Installation= | =Installation= | ||
| Line 40: | Line 42: | ||
</pre> | </pre> | ||
This will create a subdirectory <tt>obj</tt> with executables for each of the compilers supported under $G. Unless you have a compelling reason against doing so, your best bet is to stick with GCC4. Therefore, to execute, run <code>./obj/roi_select-gcc4</code>. | This will create a subdirectory <tt>obj</tt> with executables for each of the compilers supported under $G. Unless you have a compelling reason against doing so, your best bet is to stick with GCC4. Therefore, to execute, run <code>./obj/roi_select-gcc4</code>. | ||
=Using roi_select= | =Using roi_select= | ||
| Line 51: | Line 54: | ||
! {{Hl2}} | Explanation | ! {{Hl2}} | Explanation | ||
|- | |- | ||
| -i || filename[string] || | | -i || filename [string] || | ||
The source file that contains the tracks that are to be selected. | The source file that contains the tracks that are to be selected. | ||
* Supported formats are .trk, .data, and .ccf. | * Supported formats are .trk, .data, and .ccf. | ||
* It is recommended that you use the absolute path to the file. | * It is recommended that you use the absolute path to the file. | ||
|- | |- | ||
| -o || filename[string] || | | -o || filename [string] || | ||
The output file, where the selected streamlines are to be written. | The output file, where the selected streamlines are to be written. | ||
* Supported formats are .trk, .data, and .ccf. | * Supported formats are .trk, .data, and .ccf. | ||
* It is recommended that you use the absolute path to the file. | * It is recommended that you use the absolute path to the file. | ||
|- | |- | ||
| -e || expression[string] || [[#The_ROI_expression|See explanation below]] | | -e || expression [string] || [[#The_ROI_expression|See explanation below]] | ||
|} | |} | ||
| Line 72: | Line 75: | ||
|- | |- | ||
| -roi_aa || | | -roi_aa || | ||
name[string] <br /> | name [string] <br /> | ||
x1[double] <br /> | x1 [double] <br /> | ||
y1[double] <br /> | y1 [double] <br /> | ||
z1[double] <br /> | z1 [double] <br /> | ||
x2[double] <br /> | x2 [double] <br /> | ||
y2[double] <br /> | y2 [double] <br /> | ||
z2[double] | z2 [double] | ||
|| | || | ||
The first argument is the name by which you refer to this ROI in the expression. <br /> | The first argument is the name by which you refer to this ROI in the expression. <br /> | ||
| Line 84: | Line 87: | ||
|- | |- | ||
| -roi_nifti || | | -roi_nifti || | ||
name[string] <br /> | name [string] <br /> | ||
filename[string] <br /> | filename [string] <br /> | ||
region[int] | region [int] | ||
|| | || | ||
The first argument is the name by which you refer to this ROI in the expression. <br /> | The first argument is the name by which you refer to this ROI in the expression. <br /> | ||
| Line 107: | Line 110: | ||
===NIfTI Bitmap ROI=== | ===NIfTI Bitmap ROI=== | ||
'''NIfTI Bitmap ROIs''' are defined in [[NIfTI]] files. They use only three of the available dimensions and, for every voxel, store a 0 or a positive integer. A voxel with a zero value is not in any ROI. A voxel with value 1 is in ROI 1, value 2 means ROI 2, and so on. Therefore, a definition of a NIfTI Bitmap ROI consists of a filename and an integer — the particular ROI you are interested. If you want to select multiple regions from a single file, you will have to define them as separate ROIs. | '''NIfTI Bitmap ROIs''' are defined in [[NIfTI]] files. They use only three of the available dimensions and, for every voxel, store a 0 or a positive integer. A voxel with a zero value is not in any ROI. A voxel with value 1 is in ROI 1, value 2 means ROI 2, and so on. Therefore, a definition of a NIfTI Bitmap ROI consists of a filename and an integer — the particular ROI you are interested. If you want to select multiple regions from a single file, you will have to define them as separate ROIs. | ||
==The ROI expression== | ==The ROI expression== | ||
Revision as of 17:36, 2 July 2010
The Regions of Interest Selection Tool (roi_select) is a command-line program that, when provided with a collection of curves, selects and outputs only the subset that passes through a particular region (or regions) of interest.
Introduction
After we run DTI data through a pipeline (whether our in-house one or a third-party one, such as DTK), we are left with data for hundreds of thousands of streamlines. Often times, however, we are interested in only a small subset of that data. We may be working with a specific structure in the brain and would like to see a visualization of only that structure. Or we may be interested in a particular region and would like to compute statistics about the streamlines that pass through that region. Perhaps we want to see how many streamlines connect two distinct regions and what paths they take. The roi_select tool can help achieve these goals.
Installation
Dependencies
roi_select has two dependencies:
- libcurvecollection — for file I/O and in-memory data handling
- gg/args — for command-line argument processing
Both are located under $G. You will first need to set up your sandbox. (You may be able to do it automatically.)
To install libcurvecollection:
cd $G/common/libcurvecollection make all make install
To install gg_args:
cd $G/common/utility make all make install cd $G/common/gg make all make install
(see more information)
Installing roi_select
To install roi_select:
cd $G/project/brain/roi/select make all
This will create a subdirectory obj with executables for each of the compilers supported under $G. Unless you have a compelling reason against doing so, your best bet is to stick with GCC4. Therefore, to execute, run ./obj/roi_select-gcc4.
Using roi_select
After installing the dependencies and compiling the code, you can run roi_select by executing $G/project/brain/roi/select/roi_select-gcc4 with the following command-line arguments:
Required arguments
| Flag | Arguments | Explanation |
|---|---|---|
| -i | filename [string] |
The source file that contains the tracks that are to be selected.
|
| -o | filename [string] |
The output file, where the selected streamlines are to be written.
|
| -e | expression [string] | See explanation below |
At least one of these arguments is required
| Flag | Arguments | Explanation |
|---|---|---|
| -roi_aa |
name [string] |
The first argument is the name by which you refer to this ROI in the expression. |
| -roi_nifti |
name [string] |
The first argument is the name by which you refer to this ROI in the expression. |
ROI types
Axis-Aligned ROI
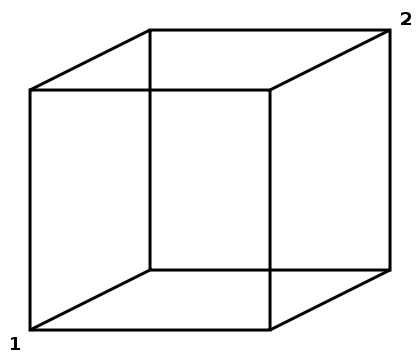
You can think of an Axis-Aligned ROI as a box placed within the image volume — that is axis-aligned, of course. A streamline is said to pass through an Axis-Aligned ROI when any of the line's vertices is located within the bounds of this box or when a line segment connecting any two adjacent vertices passes through this box.
The box that is an Axis-Aligned ROI is defined by two corners: the front bottom left and the back top right. These are labeled as corners 1 and 2 in the following diagram:
Practically speaking, we check that x1<x2, y1<y2, and z1<z2.
The coordinates of the vertices are in millimeters and must be in the same coordinate system (have the same origin) as the streamline vertices in the input file.
NIfTI Bitmap ROI
NIfTI Bitmap ROIs are defined in NIfTI files. They use only three of the available dimensions and, for every voxel, store a 0 or a positive integer. A voxel with a zero value is not in any ROI. A voxel with value 1 is in ROI 1, value 2 means ROI 2, and so on. Therefore, a definition of a NIfTI Bitmap ROI consists of a filename and an integer — the particular ROI you are interested. If you want to select multiple regions from a single file, you will have to define them as separate ROIs.